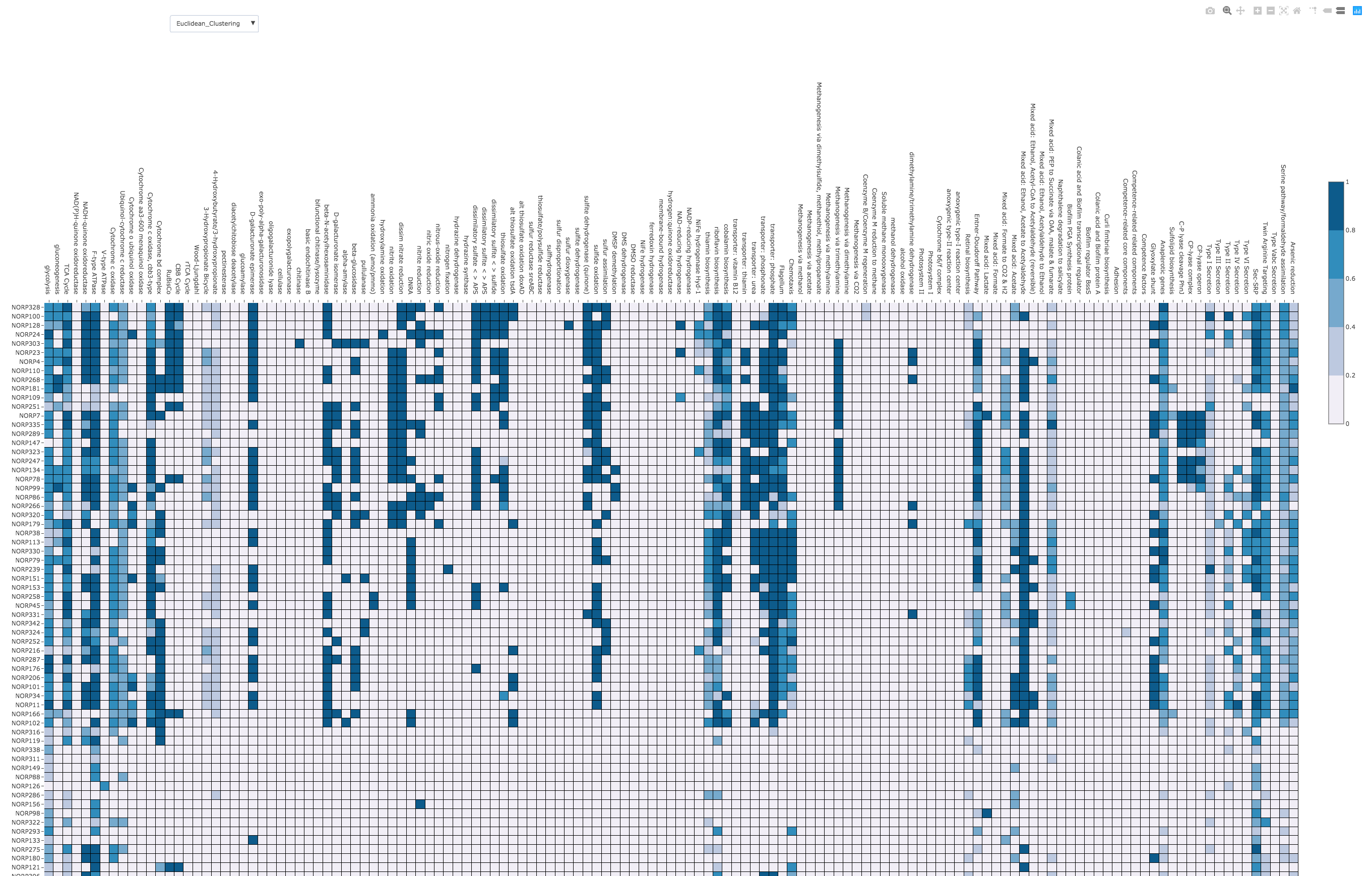
KEGGDecoder¶
GitHub repository: speeding-up-science-workshops/KEGGDecoder-heatmap/
Summary¶
- Designed to parse and visualize a KEGG-Koala output to determine the completeness of biogeochemically-relevant metabolic pathways
- metabolic pathways are hand-curated to select for informative markers of predicted function
- The immediate input for KEGGDecoder is a tab-delimited KEGG-Koala output file that consists of a formatted gene ID and a correspoding KEGG Ontology assignment
- Options for generating a KEGG-Koala output include:
- KEGG Ontology assignment requires a protein fasta file for submission and can be generated by tools such as:
- Prodigal
- MetaSanity - which can generate both a protein FASTA file and a KEGG-Koala output for a set of genomes
Authors¶
- Author Name, github id (and ORCID if you have it)
- Benjamin Tully, bjtully, 0000-0002-9384-7635
- Taylor Reiter, taylorreiter, 0000-0002-7388-421X
- Luiz Irber, luizirber, 0000-0003-4371-9659
- Roth Conrad, rotheconrad, 0000-0001-8155-8441
- Jay Osvatic, osvatic, 0000-0002-7765-0058
- Chris Neely, cjnelly10, 0000-0002-2620-8948
- Marisa Lim,
- Jason Fell, jfell13, 0000-0001-6680-2936